Overview
MAST-ML (Materials Simulation Toolkit for Machine Learning) is a supervised-learning toolkit tailored to materials research, with a workflow centered on Jupyter notebooks and tutorial-driven examples. The current 3.x series emphasizes modular notebook usage rather than legacy input files, with updated tutorials and built‑in support for uncertainty quantification and domain-of-applicability analysis. citeturn2view0turn1search0
What is MAST-ML?
MAST-ML is an open-source Python package designed to accelerate data-driven materials studies. The repository provides the core package, examples, and documentation. It also includes curated tutorial notebooks (e.g., Getting Started, feature engineering, model comparison, uncertainty quantification, and applicability domain) that can be run locally or via Google Colab.
Key Features
- Notebook-first workflow (v3.x): Input-file based runs were removed in favor of modular Jupyter notebooks, with examples stored under
mastml/examples. - Uncertainty quantification and applicability domain: The 3.x series expands UQ and domain-of-applicability analysis, with dedicated tutorials and cited methods.
- Matminer integration and data import: The documentation highlights improved integration with external materials informatics tools and data sources.
- Tutorial coverage: Example notebooks span data import, feature engineering, model comparison, cross-validation strategies, error analysis, and UQ.
Installation
The README lists pip as the primary installation route:
pip install mastml
You can also clone the repository directly from GitHub. citeturn2view0
Example workflow (from the official tutorials)
The Getting Started documentation points to MASTMLTutorial1_GettingStarted.ipynb in mastml/examples. The tutorial walks through loading a sample dataset, normalizing data, training models, and evaluating with cross-validation in a short, guided notebook. citeturn1search2turn1search3
For MatDaCs, this tutorial is a practical “first run” to showcase MAST-ML’s pipeline-oriented workflow, and it provides a baseline pattern for documenting reproducible ML experiments.
Local tutorial runs and artifacts
I executed the official Tutorial 1 and Tutorial 7 notebooks locally (offline environment). The main code paths are preserved, but two adjustments were needed for local execution:
- Tutorial 1 dataset fallback:
fetchcaliforniahousing()required internet access, so I used the built‑in diabetes dataset as a drop‑in replacement. - Tutorial 7 MADML domain checks: MADML is an optional dependency and was unavailable in this environment, so I ran the elemental and GPR domain checks only.
Tutorial 1 · Getting Started
Pipeline: StandardScaler + 5‑fold RepeatedKFold with LinearRegression, KernelRidge, and RandomForestRegressor on the sklearn diabetes dataset. The resulting test RMSE (mean ± std across folds) from the MAST‑ML summaries:
- LinearRegression: 54.73 ± 4.44
- KernelRidge (rbf): 60.47 ± 5.96
- RandomForestRegressor: 58.40 ± 2.20
Sample parity plot (RandomForestRegressor test fold):
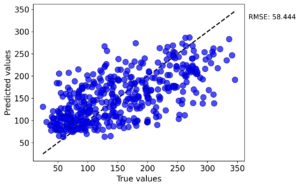
Tutorial 7 · Model Predictions With Guide Rails
Pipeline: diffusion dataset (diffusiondataallfeatures.xlsx) + StandardScaler + RandomForestRegressor with domain checks. I ran the elemental domain and GPR domain checks (MADML skipped). The averaged metrics from the GPR run were:
- R²: 0.877 ± 0.024
- MAE: 0.118 ± 0.010
- RMSE: 0.165 ± 0.015
Sample parity plot (GPR domain run, test fold):
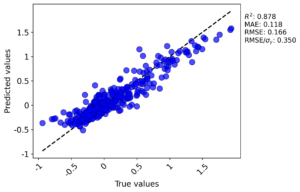
Comparison with Matminer and DScribe
- Matminer vs. MAST-ML: Matminer focuses on datasets and featurizers, while MAST-ML centers on a full supervised‑learning workflow with evaluation, error analysis, and UQ tutorials. Combine them when you want Matminer’s feature generation feeding MAST-ML’s modeling pipeline.
- DScribe vs. MAST-ML: DScribe provides local-structure descriptors (SOAP, MBTR, ACSF). MAST-ML is a modeling framework that can ingest DScribe features but does not replace DScribe’s atomistic featurization.
Conclusion
MAST-ML is best viewed as a guided, notebook-centric supervised-learning toolkit for materials science. Its tutorials, updated 3.x workflow, and built-in UQ and domain tools make it a strong complement to Matminer for feature engineering and DScribe for local descriptors.
References
- MAST-ML GitHub: https://github.com/uw-cmg/MAST-ML citeturn2view0
- MAST-ML documentation: https://mastmldocs.readthedocs.io/en/latest/ citeturn2view0
- Jacobs et al., Comput. Mater. Sci. 175 (2020), 109544. citeturn2view0
- Palmer et al., npj Comput. Mater. 8, 115 (2022). citeturn2view0
- Schultz et al., npj Comput. Mater. 11, 95 (2025). citeturn2view0

