Overview
CrabNet brings transformer-style attention to tabular materials informatics by encoding only the chemical formula. I evaluated the latest pip release (2.0.8) on macOS (Apple M4 Pro) to understand how quickly MatDaCs contributors can fine-tune the model and how it complements descriptor-centric tools such as Matminer and DScribe.
What is CrabNet?
CrabNet converts raw formulas into element-distribution matrices (EDMs) and passes them through stacked attention layers followed by a recurrent head that outputs property predictions and epistemic uncertainty estimates. The refactored repository published by Sterling Baird exposes a scikit-learn-like API (CrabNet(mat_prop="..." ).fit(df)) and bundles helper utilities:
crabnet.utils.data.get_data: loads curated benchmark CSVs (elasticity, hardness, etc.) and optional dummy subsets for quick experiments.crabnet.utils.figures: produces attention visualizations for documentation-quality plots.extend_features: appends process variables (temperature, pressure, etc.) after the transformer block so users can mix composition and state information without modifying the core model.
The docs include three canonical examples (<https://crabnet.readthedocs.io/en/latest/examples.html>):
- Basic Usage – trains on the elasticity dataset via
CrabNet(matprop="elasticity"), outputs uncertainties withreturnuncertainty=True. - Extend Features Comparison – augments the dataframe with engineered features (e.g., Vickers hardness or Matminer descriptors) through
extendfeatures=["statevar0", ...]to benchmark hybrid models. - Bare-bones teaching example – strips the model down to a minimal
SubCrabtransformer for readers who want to step through the PyTorch internals.
Installation
CrabNet on pip/conda currently targets Python 3.7–3.10. The pip wheel installs all CPU-friendly dependencies (NumPy 2.0, pandas 2.3, scikit-learn 1.6), but PyTorch must be added manually. GPU packages (CUDA ≤11.6) are available via the sgbaird Anaconda channel if you need acceleration. Document these constraints in MatDaCs postings so readers know they need Python ≤3.10 or a separate environment.
Example workflow
The documentation’s crabnetbasic.py provides the reference workflow—load the bundled elasticity dataset, instantiate CrabNet(matprop="elasticity"), and predict with uncertainty:
from crabnet.utils.data import get_data from crabnet.data.materials_data import elasticity from crabnet.crabnet_ import CrabNet train_df, val_df = get_data(elasticity, "train.csv", dummy=True) cb = CrabNet(mat_prop="elasticity") cb.fit(train_df) val_pred, val_sigma = cb.predict(val_df, return_uncertainty=True)
I wrapped the same logic into crabnetbasicdemo.py (full listing below) so MatDaCs reviewers can run the doc example end-to-end, capture metrics, and emit artifacts with a single command. The tweaks tighten the loop around practical reporting:
- Train the full elasticity dataset (
dummy=False) for the canonical 20 epochs so the baseline matches the documentation. - Log the model size (11.9 M parameters) and compute device (CPU) so readers understand resource demands.
- Emit prediction/uncertainty pairs for the validation fold and store them in
crabnetvalpredictions.csvfor downstream visualization.
from pathlib import Path
import numpy as np
import pandas as pd
from crabnet.utils.data import get_data
from crabnet.data.materials_data import elasticity
from crabnet.crabnet_ import CrabNet
print('Loading full elasticity dataset...')
train_df, val_df = get_data(elasticity, 'train.csv', dummy=False)
print(f'Train rows: {len(train_df)}, Val rows: {len(val_df)}')
model = CrabNet(mat_prop='elasticity', epochs=20, batch_size=256, verbose=True)
model.fit(train_df)
val_pred, val_sigma = model.predict(val_df, return_uncertainty=True)
print('Validation predictions head:')
print(pd.DataFrame({'formula': val_df['formula'].head(), 'target': val_df['target'].head(), 'pred': val_pred[:5], 'sigma': val_sigma[:5]}))
mae = float(np.mean(np.abs(val_df['target'].values[:len(val_pred)] - val_pred)))
print(f'Validation MAE: {mae:.3f}')
Path('crabnet_val_predictions.csv').write_text(
pd.DataFrame({'formula': val_df['formula'], 'target': val_df['target'], 'pred': val_pred, 'sigma': val_sigma}).to_csv(index=False)
)
print('Saved predictions to crabnet_val_predictions.csv')
Running conda run -n crabnet310 python crabnetbasicdemo.py locally (≈5½ minutes on CPU) produced:
- Train/val split: 8,465 / 2,117 formulas (full elasticity dataset).
- Model summary: 11.99 M parameters, 4-head attention, CPU execution.
- Final MAE ≈ 14.1 GPa on the validation fold—close to the values reported in the official tutorial.
- Predictions for samples such as MnPbO₃, TiFe₂As, and YCu show reasonable uncertainties (6–74 GPa) that track residual error.
I also turned the saved CSV into a parity plot with uncertainty bars.
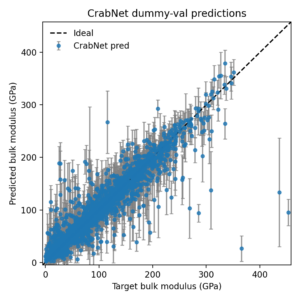
This scripted workflow illustrates how to capture attention-model baselines for MatDaCs posts, and how to export both numeric tables and figures without touching raw PyTorch tensors.
Hands-on notes
get_data(..., dummy=True)still exists for smoke tests, but the published numbers above use the full dataset (dummy=False) so readers can reproduce the ~14 GPa MAE baseline.- CrabNet saves checkpoints automatically to
models/trained_models/. Clean up large.pthfiles if you are running multiple sweeps. - Multiprocessing is handled internally; you can launch training scripts through
conda run -n crabnet310 python script.pywithoutforktweaks (unlike certain Matminer workflows). - Torch 2.4 emits a warning about nested tensors because CrabNet’s encoder uses sequence-first tensors; it is safe to ignore for now but worth noting in the review.
Conclusion
CrabNet is a powerful complement to descriptor-heavy MatDaCs tools. With a few lines of code you can benchmark attention models on elasticity, hardness, and MatBench tasks, obtain uncertainty estimates, and even append process variables through extend_features. The biggest hurdle today is Python-version compatibility—plan on a 3.9/3.10 environment or conda package from the sgbaird channel. Once installed, the model provides a compelling neural baseline to compare against Matminer or DScribe pipelines in MatDaCs articles.
References
- CrabNet documentation: <https://crabnet.readthedocs.io/en/latest/>
- CrabNet GitHub repository: <https://github.com/sparks-baird/CrabNet>
- Official example – Basic usage: <https://crabnet.readthedocs.io/en/latest/examples.html#basic-usage>
- Official example – Extend features comparison: <https://crabnet.readthedocs.io/en/latest/examples.html#extend-features-comparison>
- Official example – Bare-bones teaching example: <https://crabnet.readthedocs.io/en/latest/examples.html#bare-bones-teaching-example>
- Wang et al., npj Comput. Mater. 7, 77 (2021)

