Overview
DScribe is a Python package that accelerates the generation of atomistic descriptors for data-driven materials science. In this review I replicated the official tutorials for Cu(111) adsorption-site clustering and molecular similarity analysis to evaluate DScribe’s capabilities and usability.
What is DScribe?
Developed by SINGROUP at Aalto University, DScribe provides SOAP, MBTR, Coulomb matrix, and other descriptors for representing local atomic environments. The library integrates tightly with ASE and NumPy and is released under the Apache License 2.0.
Key Features
- Supports multiple descriptors (SOAP, MBTR, Coulomb matrix, ACSF, etc.)
- Natively interoperates with ASE
Atomsobjects and NumPy arrays - Handles batch/parallel computation, making 10k-point descriptor jobs practical on a laptop
- Includes scikit-learn compatible kernels (AverageKernel, REMatchKernel) for similarity analysis or downstream ML pipelines
Installation
DScribe installs from PyPI; dependencies such as sparse and numba are pulled automatically.
python3 -m pip install --user dscribe ase scikit-learn matplotlib
Example 1: Cu(111) adsorption-site clustering
The DScribe documentation shows how a SOAP descriptor grid above a surface can be clustered to pick representative adsorption sites instead of brute-force sampling. Following that recipe, I scanned a plane 1 Å above Cu(111), created descriptors for 10,000 positions, and ran MiniBatchKMeans to compress the search space. The job finishes in seconds and uncovers symmetry-equivalent regions automatically.
import numpy as np
import ase.build
from dscribe.descriptors import SOAP
from sklearn.cluster import MiniBatchKMeans
# Build a Cu(111) slab
system = ase.build.fcc111("Cu", (4, 4, 4), a=3.597, vacuum=10.0, periodic=True)
# SOAP descriptor setup
soap = SOAP(
species=["Cu"],
r_cut=5.5,
sigma=0.1,
n_max=12,
l_max=12,
periodic=True,
weighting={"function": "poly", "r0": 12, "m": 2, "c": 1, "d": 1},
)
# Sample a 2D grid 1 Å above the surface
cell = system.get_cell()
top = (system.get_positions()[:, 2].max() + 1.0) / cell[2, 2]
grid = np.linspace(0.0, 1.0, 100)
x, y, z = np.meshgrid(grid, grid, [top])
positions = cell.cartesian_positions(np.vstack([x.ravel(), y.ravel(), z.ravel()]).T)
D = soap.create(system, positions)
# Cluster adsorption sites
model = MiniBatchKMeans(n_clusters=10, batch_size=1024, random_state=42, n_init=5)
labels = model.fit_predict(D)
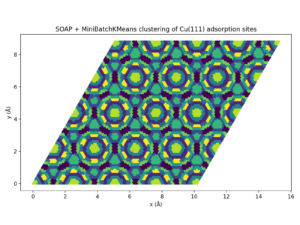
SOAP + MiniBatchKMeans clustering of Cu(111) adsorption sites
Example 2: Molecular similarity with SOAP kernels
The DScribe documentation’s “Similarity Analysis” section demonstrates how SOAP environments can be fed into Average and REMatch kernels to quantify molecular similarity with minimal code. Building on that narrative, I evaluated H2O, H2O2, NH3, and CH4, letting REMatch align local environments while the average kernel provides a quick global score. After normalizing the descriptors, both linear and RBF similarity matrices drop out ready for plotting.
from dscribe.descriptors import SOAP from dscribe.kernels import AverageKernel, REMatchKernel from ase.build import molecule from sklearn.preprocessing import normalize names = ["H2O", "H2O2", "NH3", "CH4"] structures = [molecule(name) for name in names] soap = SOAP(species=["H", "C", "N", "O"], r_cut=5.0, sigma=0.2, n_max=2, l_max=2, periodic=False) features = [soap.create(struct) for struct in structures] features_norm = [normalize(f) for f in features] avg_kernel = AverageKernel(metric="linear").create(features) re_kernel = REMatchKernel(metric="rbf", gamma=1.0, alpha=1.0).create(features_norm)
Application Areas
- Catalysis & surface science: Clustering adsorption sites, selecting active motifs
- Solid-state screening: SOAP/MBTR descriptors for structure comparison and ML-driven discovery
- Molecular design: REMatch-based similarity scoring for candidates in cheminformatics
- ML model building: Feeding descriptors into regression/classification/clustering models to predict properties
Comparison with Similar Tools
- ASE alone: Limited built-in descriptors; DScribe offers extensive feature sets without custom coding.
- quippy/libatoms: Powerful for GAP potentials, but DScribe’s Python API and documentation make general ML workflows easier.
- Matminer: Great for bulk/material dataset descriptors, but DScribe excels at local environment features and efficiency.
Hands-on Notes
- Ease of use: Tutorials run almost verbatim; Friendly for ASE/NumPy users.
- Performance: Computing 10,000 SOAP vectors of length 1,014 took ~8 seconds on a notebook (Apple M4 Pro).
- Numerical stability: Set
randomstate/ninitfor reproducible MiniBatchKMeans; normalize inputs before REMatchKernel to avoid numerical issues. - Workflow: Persisting
D.npyandr.npyenables quick handoff to visualization, optimization, or ML tasks.
Conclusion
DScribe is a versatile descriptor engine that streamlines machine-learning workflows in materials science. It covers both adsorption-site analysis and molecular similarity tasks out of the box and integrates smoothly with the Python ML stack, making it a strong recommendation for MatDaCs users exploring descriptor-based approaches.
References
- DScribe documentation: <https://singroup.github.io/dscribe/latest/>
- Tutorial (Clustering): <https://singroup.github.io/dscribe/latest/tutorials/machine_learning/clustering.html>
- Tutorial (Similarity Analysis): <https://singroup.github.io/dscribe/latest/tutorials/similarity_analysis/kernels.html>
- GitHub repository: <https://github.com/SINGROUP/dscribe

