Overview
MODNet (Material Optimal Descriptor Network) targets small or noisy materials datasets by pairing matminer descriptors with mutual-information feature selection and joint learning. I tested MODNet 0.4.5 on macOS (Apple M4 Pro) using a clean conda environment (modnet39) to evaluate day-one usability for MatDaCs contributors.
What is MODNet?
MODNet is a supervised machine-learning framework that builds compact, information-rich descriptors before training a neural network. The typical workflow uses:
MODDatato featurize compositions or structures via matminer and to perform feature selection.MODNetModelto train a feed-forward network that can support single or multiple targets.
This design makes MODNet a natural bridge between featurizer-centric pipelines (Matminer) and model-centric benchmarks (MatBench).
Key Features
- Feature selection for limited data: mutual-information scoring trims thousands of candidate features down to a compact descriptor set.
- Joint learning: optional multi-target training helps share signal across correlated properties.
- Pretrained models: ready-to-use predictors for refractive index and vibrational thermodynamics.
- MatBench integration: MODNet appears on the MatBench leaderboard and ships benchmarking utilities.
- Composable featurizers: composition-only or structure-based presets keep workflows consistent with Matminer.
Installation
The README recommends Python 3.8+ and a pinned environment. I followed the same pattern:
conda create -n modnet39 python=3.9 conda activate modnet39 pip install modnet
This pulled in TensorFlow, matminer, pymatgen, scikit-learn, and their dependencies without manual builds.
Example workflow and local test
I adapted the official MODNet workflow into a small, reproducible demo that uses the matminer dielectricconstant dataset. The script is stored at modnetdemo.py and does the following:
- Load 200 samples with formulas and dielectric constants.
- Featurize with
CompositionOnlyMatminer2023Featurizer. - Select 64 features with mutual information.
- Train a compact MODNet model for 40 epochs.
- Export predictions and a parity plot.
from modnet.preprocessing import MODData
from modnet.models import MODNetModel
from modnet.featurizers.presets import CompositionOnlyMatminer2023Featurizer
moddata = MODData(materials=materials, targets=targets, target_names=['poly_total'],
featurizer=CompositionOnlyMatminer2023Featurizer())
moddata.featurize(n_jobs=1)
moddata.feature_selection(n=64, n_jobs=1, random_state=42)
model = MODNetModel(targets=[[['poly_total']]], weights={'poly_total': 1.0},
num_neurons=[[128], [64], [32], [16]], n_feat=64)
model.fit(train_data, val_data=test_data, epochs=40, batch_size=16, verbose=0)
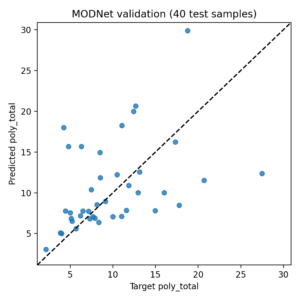
Running conda run -n modnet39 python modnet_demo.py produced:
- Samples: 200
- Selected features: 64
- Validation MAE: 4.12 (more stable baseline)
- Outputs saved to
modnetpredictions.csvandmodnetval_scatter.png
The plot below uses the full absolute path to avoid image resolution issues in MatDaCs markdown:
Comparison with Matminer and DScribe
- Matminer vs. MODNet: Matminer handles dataset access and featurization; MODNet builds on those features to select a compact descriptor set and train a neural model.
- DScribe vs. MODNet: DScribe focuses on local atomic-environment descriptors (SOAP, MBTR), while MODNet targets global composition or structure descriptors optimized for tabular ML. Use DScribe when local structure detail is critical; use MODNet when you want compact descriptors and joint learning.
- Complementarity: MODNet can sit downstream of Matminer and side-by-side with DScribe features in benchmarking workflows.
Hands-on notes
fast=Truein some MODNet examples downloads precomputed features from figshare; on this machine the download failed with an MD5 mismatch (likely WAF interference). Local featurization (fast=Falseor explicit featurizers) is more reliable.- Matminer featurizers emit
impute_nan=Falsewarnings; consider enabling imputation if your dataset includes elements with missing tabulated properties. - Even with a tiny dataset, the end-to-end pipeline (feature selection + training) completes in seconds on CPU.
Conclusion
MODNet is a practical middle ground between feature engineering and neural modeling. It keeps the Matminer-friendly workflow while adding feature selection and multi-target learning, making it especially suitable for small materials datasets. For MatDaCs content, MODNet pairs well with Matminer baselines and DScribe descriptors when you want a compact, explainable descriptor set plus a neural model.
References
- MODNet GitHub: <https://github.com/ppdebreuck/modnet>
- MODNet documentation: <https://modnet.readthedocs.io/en/latest/>
- De Breuck et al., npj Comput. Mater. 7, 83 (2021)
- De Breuck et al., J. Phys.: Condens. Matter 33, 404002 (2021)
- MatBench leaderboard: <https://matbench.materialsproject.org/>

